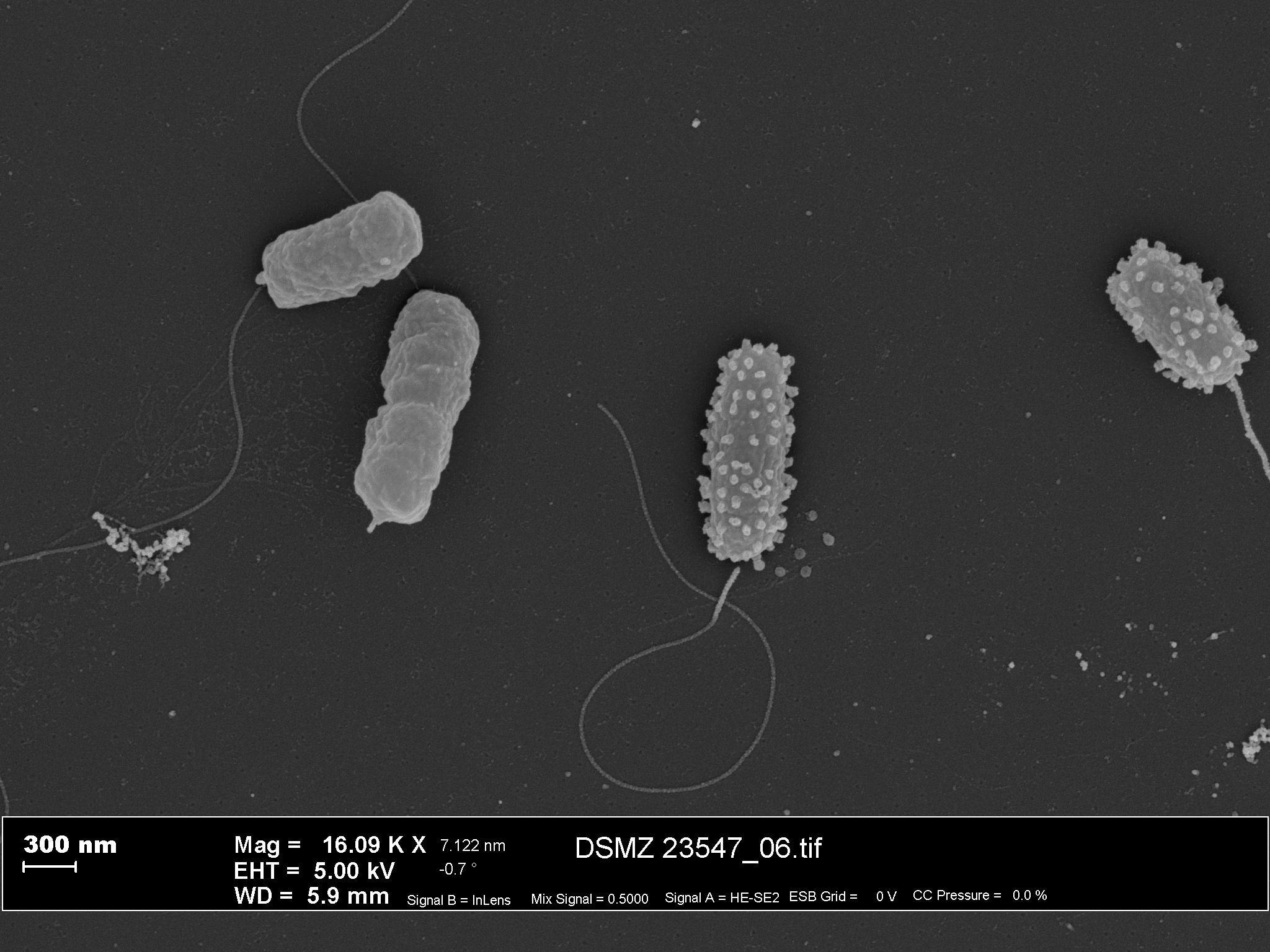
News
last update: 13.09.2024-
BacDive Feature Update
BacDive's new Special Collections assemble strains that are part of the same research project or collection. This provides users with better access and makes these data sets more visible. You can find all Special Collections on a dedicated new page at https://bacdive.dsmz.de/special-collections. Have fun exploring!

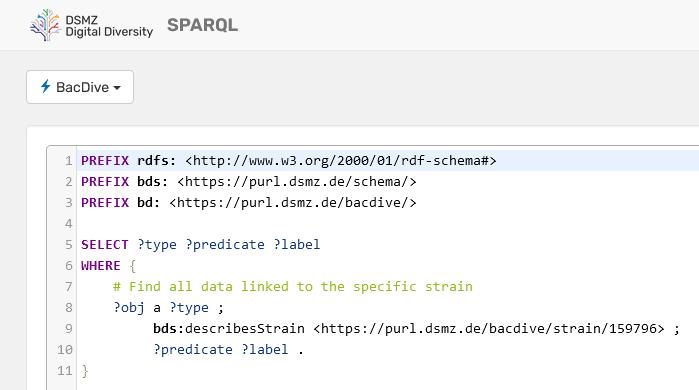
We are also very happy to present the new BacDive knowledge graph, which contains more than 16.5 million triples representing so far 26 of the most important BacDive entities. It can be accessed through a SPARQL endpoint (https://sparql.dsmz.de/bacdive) that enables direct and standardized searches of BacDive data and also allows for federated queries spanning multiple data sources.
-
BacDive Mini Update
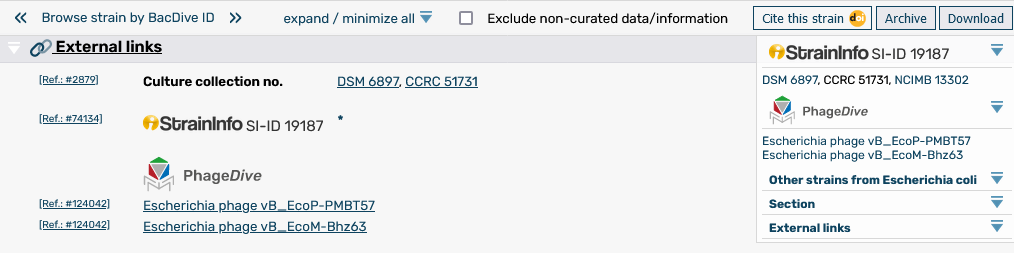
BacDive pages now contain links to the new PhageDive database for information on prokaryotic viruses..
5308 new 16S rRNA gene sequences were automatically matched to BacDive strains.
Additionally, several small bugs were fixed to improve the BacDive user experience.
-
New BacDive Release
We are happy to announce that the traditional Christmas BacDive release is now online.
This update features the integration of a large set of phenotypic and metabolic data generously shared by the Biological Resource Center (CRBIP) of the Institut Pasteur. BacDive now provides data on 97,334 strains, 4080 of which were newly added. The number of total filled data fields grew by almost 30% to 2,675,988 entries.
A careful curation of the genome sequences listed in BacDive resulted in the removal of links to low-quality assemblies and the consolidation of listings of single contigs of WGS projects in assembly accession. The number of sequence entries therefore decreased, while the quality of displayed genomes increased.
BacDive strain pages now contain links to the newly revived StrainInfo database for strain identity information and the resolution of strain identifiers.
Genome-based predictions from the Diaspora project can now be accessed using the API using an optional parameter.
We want to thank all of our users who have sent in suggestions and corrections over the last year, many of which were accepted and become visible with this release.
BacDive Section Release 2021 Release 2022 Release 2023 2022-2023 increase Name and taxonomic classification* 132,839 137,332 154,525 11% Morphology* 42,756 62,766 74,683 16% Culture and growth conditions* 157,622 159,833 193,351 17% Physiology and metabolism* 1,063,988 1,112,931 1,691,087 34% Isolation, sampling and environmental information* 156,065 175,521 187,071 6% Application and interaction* 22,077 23,221 36,338 36% Sequence data* 106,070 122,022 109,572 -11% References* 68,145 128,540 229,361 44% overall entries* 1,749,562 1,922,166 2,675,988 28% BacDive is ELIXIR Core Data Resource
We are proud that after an extensive review and evaluation process, BacDive has been selected as a new ELIXIR Core Data Resource, joining a list of now 32 renowned European life science data resources that are important to biological and biomedical research and long-term preservation of biological data.

-
New BacDive Release
BacDive steps into the AI world!
Using support vector machine and deep learning algorithms on genomes of 9023 bacterial strains, we added high quality predicted data to our database including data on:
- - Gram staining
- - Oxygen tolerance
- - Spore formation
- - Motility
- - Halophily
- - Thermophily
- - Acidophily
Check some of our strains with predicted data: Acetobacter pasteurianus, Alicyclobacillus acidoterrestris
Prediction data is not available yet in the BacDive API. It will be soon available on demand.

-
New BacDive Release
A new BacDive release is online, providing now data on 93,254 strains!
This update includes a major update for data from DSMZ database, including 637 new strains.
We also integrated comprehensive data on 267 strains for the Wink compendium of Actinobacteria, which now comprises data on 1576 strains and is now finalized!
An exiting new feature are our new taxonmaps: for 15.655 strains we could match 16S sequence data with the database MicrobeAtlas and now provide integrated overviews over the global distribution of a taxon.
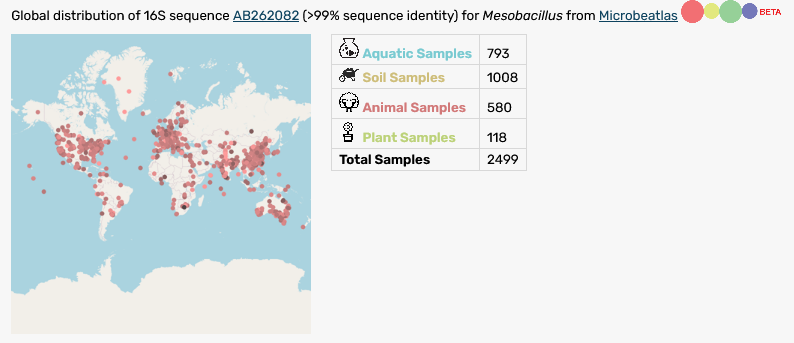
Find the overall statistics in the table (below).
BacDive Section Release 2020 Release 2021 Release 2022 2021-2022 increase Name and taxonomic classification* 110,587 132,839 137,332 3% Morphology* 36,689 42,756 62,766 47% Culture and growth conditions* 136,586 157,622 159,833 1% Physiology and metabolism* 332,303 1,063,988 1,112,931 5% Isolation, sampling and environmental information* 143,496 156,065 175,521 12% Application and interaction* 21,710 22,077 23,221 5% Sequence data* 295,734 106,070 122,022 15% References* 66,571 68,145 128,540 89% overall entries* 1,143,676 1,749,562 1,922,166 10% -
BacDive selected as Global Core Biodata Resource
The BacDive database of the DSMZ was selected as one of 37 databases worldwide as a Global Core Biodata Resource.
In a rigorous selection process, the Global Biodata Coalition chose such resources that are considered critical for global research in the fields of life sciences and biomedicine. The BacDive database has been developed at the DSMZ for 10 years and now joins the list of the world's most important biological databases.
See https://globalbiodata.org/global-biodata-coalition-announces-the-first-set-of-global-core-biodata-resources/ for more information.
-
New BacDive Release
A new BacDive release is online. 2604 new type strains from LPSN with their corresponding 2447 sequence accession numbers (16S) and risk assessment information have been integrated in our database.
Currently we offer metadata on 92,150 strains, including 18,710 type strains. An 16% type strains increase since the last update
Some suggestions from our users were included in this update as well. Thank you for making BacDive better!
To find even more information about our strains, we added 56,017 extra literature resources to allow you to dig even deeper into the strains of your interest.
We are also very proud of our new feature, the Smartsearch. Using a google like search now you can search our database for taxonomy, culture collection numbers, NCBI taxid, sequence accession numbers, motility, cell shape and much more.
-
New BacDive License
We have made the BacDive License even more permissive with the Creative Commons Attribution 4.0 International License.
-
New BacDive Release
A new BacDive release is online since December 2021 comprising the largest update in the history of BacDive.
Compared to the major update in 2020, the overall data content has increased by 80 %. Currently we offer metadata on 89,545 strains, including 16,105 type strains.
The major sources for this update are new data from the culture collections JCM, KCTC and UCCCB. These comprise a lot of detailed information for 14,658 strains, including 5711 new strains. From the culture collection DSMZ, data on 942 new strains were integrated.
The second source is the regular update of manually annotated information on type strains from species descriptions. With this update detailed information on 687 strains were added, so that BacDive now provides 7585 data sets from literature.
Furthermore, we decided to transcribe API® test data where possible into the according BacDive data fields. This concerns mostly enzymatic and metabolic data. Data that originate from API® test data are clearly labeled as such!
*)Denotes the distinct combination of strain, reference, data entryBacDive Section Release 2019 Release 2020 Release 2021 2020-2021 increase Name and taxonomic classification* 108,630 110,587 132,839 20% Morphology* 35,965 36,689 42,756 17% Culture and growth conditions* 132,494 136,586 157,622 15% Physiology and metabolism* 312,973 332,303 1,063,988 220% Isolation, sampling and environmental information* 138,474 143,496 156,065 9% Application and interaction* 19,774 21,710 22,077 2% Sequence data* 35,258 295,734 411,718 39% References* 65,427 66,571 68,145 2% overall entries* 848,995 1,143,676 2,055,210 80% -
New BacDive Feature Update
A new BacDive feature update is online, with the following new functionalities:
- Homepage Relaunch with new design - Modular Advanced Search.
- Modular Advanced Search.
This new search replaces the advanced search offering more powerfull searching capabilities and user friendliness. - NCBI tax ID is now searchable on the main search using the format "taxid: 1604334"
- NCBI tax ID is now searchable on the main search using the format "taxid: 1604334" -
New BacDive mailing list and API
The BacDive Webservice has been renewed to adapt to our users needs and offer a better performance and comfort.
Find the new webpage here: https://api.bacdive.dsmz.de/
We upgraded the authorisation system. With a single sign on you will be able to access all the Web services of the DSMZ including BacDive and LPSN.
With the new mailing list you can get informed about new features and about upcoming updates and changes in BacDive.
You can subscribe any time here: /mailinglist/subscribe
-
New BacDive Release
A new BacDive release is online since December 2020 comprising new data content and new features
Compared to the major update in 2019, the overall data content has increased by 35 %. Currently we offer metadata on 82,892 strains, including 14,208 type strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 2018 Release 2019 Release 2020 2019-2020 increase Name and taxonomic classification* 89,307 108,630 110,587 2% Morphology* 30,232 35,965 36,689 2% Culture and growth conditions* 111,414 132,494 136,586 2% Physiology and metabolism* 198,529 312,973 332,303 6% Isolation, sampling and environmental information* 77,883 138,474 143,496 4% Application and interaction* 19,111 19,774 21,710 10% Sequence data* 30,475 35,258 295,734 739% References* 43,210 65,427 66,571 2% overall entries* 600,161 848,995 1,143,676 35%
A major change in this update is the restructuring of the sections: Morphology & Physiology was splitted into the two sections Morgphology and Physiology & Metabolism. Molecular Biology was renamed to Sequence Data and the sequence data table was splitted into 16S and genome Sequence data. A large amount of sequence accession data was introduced by automatic annotation, which was developed within the new DiASPora-project.
A new summary called "BacPhrases" was introduced at the top of each strain detail view. Giving a fast overview over some important data about the strain within a short sentence, followed by a couple of keywords.
Manually annotated data from IJSEM species descriptions were integrated, giving a total of 6898 data sets with species description data. -
New minor BacDive Release
A new statistic tool is available: the new dashboard enables to explore BacDive data in a new interactive way. 49 different graphs within seven sections give a fast overview of the data provided in the database including all strains. For a representative overview, data can be filtered for type strain data only. By clicking on data points in the graphs queries are starting in the respective specialized search tools, e.g. Advanced Search, TAXplorer, Isolation source search, or API test finder.
-
New minor BacDive Release
-
New BacDive Release
A new BacDive release is online since December 2019 comprising new data content and new features
Compared to the major update in 2018, the overall data content has increased by 36 %. Currently we offer metadata on 81,827 strains, including 14,091 type strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2017 Release 07/2018 Release 12/2019 2017-2018 increase Name and taxonomic classification* 111,832 156,229 212,578 36% Morphology and physiology* 126,992 228,761 348,938 53% Culture and growth conditions* 77,471 119,126 132,494 11% Isolation, sampling and environmental information* 25,509 77,883 138,474 78% Application and interaction* 18,863 19,111 19,774 3% Molecular biology* 23,349 30,475 35,258 16% Strain availability* 68,853 81,373 82,538 1% overall entries* 452,869 712,958 970,054 36%
Analytical test data like Analyitcal Profile Index (API) data and fatty acid profiles were imported from the Swedish Culture Collection University of Gothenburg (CCUG). By integrating the data, the API data collection provides now 27,634 profiles for 15,357 strains and is the largest publicly available database in the world. The API database can be queried using the API test finder. The fatty acid profile database now comprises 5216 data sets.
Manually annotated data from IJSEM species descriptions were integrated, giving a total of 6640 data sets with species description data.
The Isolation source search was extended by new functions like the display of the geographic distribution on a world map and the analyses of the isolation source tags in a Krona plot.
-
New BacDive Release
A new BacDive release is online since April 2019 comprising new data content and new features
Compared to the major update in 2018, the overall data content has increased by 26 %. Currently we offer metadata on 80,584 strains, including 13,498 type strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2017 Release 07/2018 Release 04/2019 2017-2018 increase Name and taxonomic classification* 111,832 156,229 208,353 33% Morphology and physiology* 126,992 228,761 295,040 29% Culture and growth conditions* 77,471 119,126 125,775 6% Isolation, sampling and environmental information* 25,509 77,883 136,791 76% Application and interaction* 18,863 19,111 19,111 0% Molecular biology* 23,349 30,475 34,578 13% Strain availability* 68,853 81,373 81,373 0% overall entries* 452,869 712,958 901,021 26%
Data for 19,505 strains were imported from the Swedish Culture Collection University of Gothenburg (CCUG). Moreover, manually annotated data from 756 IJSEM species descriptions were integrated, giving a total of 6224 data sets with species description data. Over 1500 pictures were imported from the internal files of the DSMZ showing e.g. colony morphology on agar plates or sporulation.
DSM 20300 Corynebacterium glutamicum
DSM 14716 Cystobacter ferrugineus
-
Publication out now
The latest Publication on BacDive is now available.
BacDive in 2019: bacterial phenotypic data for High-throughput biodiversity analysis
Reimer, L. C., Vetcininova, A., Sardà Carbasse, J., Söhngen, C., Gleim, D., Ebeling, C., Overmann, J.
Nucleic Acids Research; database issue 2019.
Please cite BacDive and results obtained by the use of the BacDive database. -
New BacDive Release
A new BacDive release is online since July 2018 comprising new data content and new features
Compared to the major update in 2017, the overall data content has increased by 146 %. Currently we offer metadata on 63,669 strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2017 Release 01/2018 Release 07/2018 2017-2018 increase Name and taxonomic classification* 111,832 154,264 156,229 51 % Morphology and physiology* 126,992 198,091 228,761 485 % Culture and growth conditions* 77,471 117,181 119,126 391 % Isolation, sampling and environmental information* 25,509 35,840 77,883 247 % Application and interaction* 18,863 18,855 19,111 22 % Molecular biology* 23,349 28,279 30,475 76 % Strain availability* 68,853 80,600 81,373 22 % overall entries* 452,869 633,110 712,958 146 %
New Microbial Isolation Source tags were introduced that describe the original isolation source entry with terms from a controlled vocabulary and make them systematically searchable. Additionally, a map was implemented, that displays the source of isolation, either based on geographic coordinates (if available) or on country data.
To make strains with microbial isolation source tags searchable, the Microbial Isolation Source Search was develeoped. This tool enables the search for multiple isolation source tag combinations and allows to filter the result by additional fields like species name, country or the original isolation source entry.
-
New BacDive Release
A new BacDive release is online since January 2018 comprising new data content.
The overall data content has increased by 40 %. Currently we offer metadata on 62,683 strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2015 Release 09/2016 Release 09/2017 Release 01/2018 2017-2018 increase
Name and taxonomic classification* 106,952 108,226 111,832 154,264 38% Morphology and physiology* 39,087 54,129 126,992 198,091 56% Culture and growth conditions* 24,280 58,206 77,471 117,181 51% Isolation, sampling and environmental information* 22,448 23,077 25,509 35,840 40% Application and interaction* 15,680 15,873 18,863 18,855 0% Molecular biology* 17,668 20,029 23,349 28,279 21% Strain availability* 66,832
68,798 68,853 80,600 17% overall entries* 292,947 373,628 452,869 633,110 40%
The increase in data content originates from two new data sources:
- Data for 9,592 strains were integrated from the Collection de l’Institut Pasteur (CIP).
- Data for 4,082 strains were integrated from the publication "Hiding in Plain Sight: Mining Bacterial Species Records for Phenotypic Trait Information", which provides extracted metadata from IJSEM publications.
-
New BacDive Major Release
The new BacDive major release is online since September 2017 with some new features and new data content.
The overall data content has increased by 30 %. Currently we offer metadata on 56,420 strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2013Release 09/2014 Release 09/2015 Release 09/2016 Release 09/2017 2016-2017 increase
Name and taxonomic classification* 23,458 104,922 106,952 108,226 111,832 3% Morphology and physiology* 11,521 23,928 39,087 54,129 126,992 135% Culture and growth conditions* 21,605 23,696 24,280 58,206 77,471 33% Isolation, sampling and environmental information* 20,769 21,873 22,448 23,077 25,509 11% Application and interaction* 14,639 15,637 15,680 15,873 18,863 19% Molecular biology* 14,591 16,168 17,668 20,029 23,349 17% Strain availability* 18,459 62,888 66,832
68,798 68,853 0.1% overall entries* 125,042 269,112 292,947 373,628 452,869 30%
Besides the increase of data content, we offer some new features in the BacDive Webportal:
API test finder: The API test finder is a new tool to find matching API tests within the database.
Antibiotic susceptibility tests
Data on antibiotic susceptibility tests is now available. Inhibition zone diameters can be queried via the advanced search.
Digital Object identifier (DOI) and PDF archiving system
The PDF archive for each strain and the according DOIs can be found under "PDF archive" in the strain detail view (upper right corner).
-
Video tutorials online
BacDive now offers short video tutorials on the usage of the BacDive GUI. These tutorials deal with the topics "basic functions", "advanced search" and "export functions" and can be found in the tutorial section.
-
New BacDive Minor Release
The new BacDive minor release is online since February 2017. This update includes some smaller database changes and additional data content, including extracted data from 523 initial strain descriptions published in the International Journal of Systematic and Evolutionary Microbiology (IJSEM). This data set comprises predominantly data on the morphology and physiology of strains and in consequence the data content of this section has increased by 74 %. Due to optimization of data structures the content for Culture and growth conditions has slightly decreased. Overall the data content has increased by 11 % and BacDive now offers metadata on 55.687 strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2013Release 09/2014 Release 09/2015 Release 09/2016 Release 02/2017 2016-2017 increase
Name and taxonomic classification* 23,458 104,922 106,952 108,226 110,385 2% Morphology and physiology* 11,521 23,928 39,087 54,129 94,158 74% Culture and growth conditions* 21,605 23,696 24,280 83,496 74,024 -11% Isolation, sampling and environmental information* 20,769 21,873 22,448 23,077 24,648 7% Application and interaction* 14,639 15,637 15,680 15,873 18,766 18% Molecular biology* 14,591 16,168 17,668 20,029 22,565 13% Strain availability* 18,459 62,888 66,832
68,798 68,991 0.3% overall entries* 125,042 269,112 292,947 373,628 413,537 11%
-
Downtime due to server maintenance at 10./11.12
Due to a server maintenance, BacDive will be temporarily unavailable during the 10./11.12.
-
User satisfaction survey
BacDive is a part of the German Network for Bioinformatics Infrastructure de.NBI.
To evaluate and improve the quality of our service we need your feedback.
Please help us by participating in this short survey.
-
Training for the use of BacDive on 2nd of November 2016
In the course of the de.NBI training sessions there will be a training for the database BacDive. The training comprises an introduction, a detailed presentation of the utilization possibilities and comprehensive exercises. Please find detailed information in this flyer or at the de.NBI page.
The training is free of charge. However the participants have to cover their travelling and hotel expenses. The application is open to all though de.NBI members will be preferred.
Location:
DSMZ-Building at the HZI-Campus Braunschweig
Inhoffenstraße 7B 38124 Braunschweig
Date:
2. November 2016, 10 a.m. to 4 p.m.Please register at training@bacdive.de
-
New BacDive Major Release
The new BacDive major release is online since September 2016 with a refurbished Webportal and some new features. Additionally the data content has increased by 23%. Currently we offer metadata on 54,610 strains.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2013Release 09/2014 Release 09/2015 Release 09/2016 2015-2016 increase
Name and taxonomic classification* 23,458 104,922 106,952 108,226 4% Morphology and physiology* 11,521 23,928 39,087 54,129 28% Culture and growth conditions* 21,605 23,696 24,280 83,496 71% Isolation, sampling and environmental information* 20,769 21,873 22,448 23,077 3% Application and interaction* 14,639 15,637 15,680 15,873 1% Molecular biology* 14,591 16,168 17,668 20,029 13% Strain availability* 18,459 62,888 66,832
68,798 3% ? overall entries* 125,042 269,112 292,947 373,628 23%
Besides the increase of data content, we offer some new features in the BacDive Webportal:
-
Publication out now
The latest Publication on BacDive is now available.
BacDive - the Bacterial Diversity Metadatabase in 2016
Söhngen C., Bunk B., Podstawka A., Gleim D., Vetcininova A., Reimer L.C., Overmann J.
Nucleic Acids Res. 2015. doi: 10.1093/nar/gkv983. Epub 2015 Sep 30.
PMID: 26424852
Please cite BacDive and results obtained by the use of the BacDive database. -
New BacDive Release
The new BacDive major release is online since September 2015 with a substantially increase of content. Currently we offer metadata on 53,978 strains.
These data is derived out of 22,033 source references
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2013 Release 09/2014 Release 09/2015 2014-2015 increase 2013-2015 increase
Name and taxonomic classification* 23,458 104,922 106,952 2% 356% Morphology and physiology* 11,521 23,928 39,087 63% 239% Culture and growth conditions* 21,605 23,696 24,280 2% 12% Isolation, sampling and environmental information* 20,769 21,873 22,448 3% 8% Application and interaction* 14,639 15,637 15,680 - 7% Molecular biology* 14,591 16,168 17,668 9% 21% Strain availability* 18,459 62,888 66,832
6% 262% ? overall entries* 125,042 269,112 292,947 9% 134%
Besides the increase of data content, we offer some new features of the BacDive Webportal:
Search for species in external web resources:
»
Search for information on refering species in external web resources, e.g. BRaunschweig ENzyme DAtabase (BRENDA), Global Biodiversity Information Facility (GBIF), PANGAEA, StrainInfo Enhancement of the section molecular biology:
» The section molecular biology offers some more data fields: BacDive strain entries with International Nucleotide Sequence Database Collaboration (INSDC) accession numbers are now complemented by the brief sequence accession description and the associated ID of the NCBI Taxonomy. Additionally we annotated accession numbers of completed genomes of 654 strains.
Additional fields in the advanced search:
» In order to search for gene names, plasmids or 16S rRNA in BacDive use the advanced search to query the field sequence accession description. The BacDive advanced search is now also capable to retreive entries by a given ID of the NCBI Taxonomy.
-
Training for the use of the databases BacDive and BRENDA in October 2015
In the course of the de.NBI training sessions there will be a training for the databases BacDive and BRENDA. The goals of this training are to introduce the databases and to give detailed information on the scope, the organization, the content, and how to work with the tools. The training is open to all, but de.NBI members will be given preference.
Please find more detailed information about the program and the registration in this flyer. The training is free of charge. However the participants have to cover their traveling and hotel expenses.
Location:
Technische Universität Braunschweig, Braunschweig, Germany
Date:
13.+14. October 2015
Registration deadline:
15.09.2015 for partners of the de.NBI Network
1.10.2015 for all others
-
Assembling the microbial diversity jigsaw puzzle -5 demonstrators for data mobilization
BacDive strives for more manually annotated content derived out of publications. The limited scope of five bacterial strains have been in the focus of this mobilization pilot project. Although a biological resource might be of crucial importance for the environmental state of a certain biotope, it is still laborious to screen and compare certain properties of candidate strains.This holds also true if a strain is already deposited within a culture collection and has become a research object with published data. The information about the strain is scattered in publications and data bases around the world.
The data mobilization of five bacterial strains demonstrates the valuable and comprehensive data sources represented by culture collection data base content and the related research results and publications. For this mobilization demonstrators the taxon-related descriptive information has been manually annotated out of the referring publications and added to BacDive. In this attempt an average of 276 (180-319) data points per strain could be mobilized.
The ecological and environmental relevance of these strains is given by their participation in nitrogen and carbon cycles (significant exoenzyme activity) within their particular habitat. The detailed documented metabolic and physiologic profile of each strain indicates their significant adaptation to the soils of different climates. Hence, these strains are important biotic components of their environment.
The detailed descriptions in BacDive:
-
Beta-Tests completed
We thank all those who have tested our web services and gave us feedback on their optimization. The BacDive web services are available to all interested BacDive users. If you like to obtain an account, you simply need to register (Registration forms BacDive, PNU). -
Beta-Testers Wanted!
We are providing the new BacDive web service as beta release. All endpoints are comprehensively documented. The content can be retrieved in machine readable XML and JSON format. If you are interested in becoming a beta tester, please apply via the provided registration forms. -
Additional information on more than 1000 strains available
The Compendium of Actinobacteria is an comprehensive electronic PDF manual including the descriptions of the important bacterial calls of Actinobacteria compiled by PD Dr. Joachim Wink ( HZI-Helmholtz-Centre for Infection Research, Braunschweig). The descriptions of morphology and physiology are now available as structured database content within BacDive. Here some representative links:
-
New BacDive Release
-
Dive Major Release
The new BacDive major release is online since September 2014 with a substantially increase of content. Currently we offer metadata on 53,588 strains. These data is derived out of 21,222 source references.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2012
Release 09/2013
Release 09/2014 2013-2014 increase Name and taxonomic classification* 18,157 23,458 104,922 347% Morphology and physiology* 4,053 11,521 23,928 108% Culture and growth conditions* 20,634 21,605 23,696 10% Isolation, sampling and environmental information* 15,449 20,769 21,873 5% Application and interaction* - 14,639 15,637 7% Molecular biology* 13,609 14,591 16,168 11% Strain availability* 17,856 18,459 62,888 241% ? overall entries* 89,758 125,042 266,718 241%
Besides the increase of data content, we offer some new features of the BacDive Webportal.- The BacDive web service is available as alpha release. All endpoints are comprehensively documented. The data is available in XML and JSON format. If you are interested in becoming a beta tester, please apply via the provided registration forms.
- Linking to StrainInfo.net: BacDive now offers links to the corresponding strain passport in StrainInfo.net.
-
Publication out now
The Publication on BacDive is now available.
BacDive - the Bacterial Diversity Metadatabase
Söhngen C., Bunk B., Podstawka A., Gleim D., Overmann J.
Nucleic Acids Res. 2014 Jan 1;42(1):D592-9. doi: 10.1093/nar/gkt1058. Epub 2013 Nov 7.
PMID: 24214959
Please cite BacDive and results obtained by the use of the BacDive database. -
New BacDive Major Release
The new BacDive major release is online since September 2013 with a substantially increase of content.
*) Denotes the distinct combination of strain, reference, data entryBacDive Section Release 09/2012 Release 09/2013 2012-2013 increase Name and taxonomic classification* 18,157 23,458 29% Morphology and physiology* 4,053 11,521 184% Culture and growth conditions* 20,634 21,605 5% Isolation, sampling and environmental information* 15,449 20,769 34% Application and interaction* 0 14,639 100% Molecular biology* 13,609 14,591 7% Strain availability* 17,856 18,459 3% ? overall entries* 89,758 125,042 39%
Besides the increase of data content, we have added and enhanced some features of the BacDive Webportal.- The BacDive advanced search has been adapted and offers now large-scale queries functionality by combining more than 30 data fields for your individual comparative analyses of a multitude of strains.
- We added Help and FAQ that provide you with a quick and easy introduction to the use of the BacDive features and functionalities.