| [Ref.: #5321] |
Culture collection no. |
DSM 14365, JCM 11303, AJ 13395, CIP 107738 |
| [Ref.: #75349] |
 SI-ID 100667 SI-ID 100667
|
* |
 |
Literature: |
Only first 10 entries are displayed. Click here to see all.Click here to see only first 10 entries. |
|
Topic |
Title |
Authors |
Journal |
DOI |
Year |
|
| Phylogeny |
Analysis of the Genome and Metabolome of Marine Myxobacteria Reveals High Potential for Biosynthesis of Novel Specialized Metabolites. |
Amiri Moghaddam J, Crusemann M, Alanjary M, Harms H, Davila-Cespedes A, Blom J, Poehlein A, Ziemert N, Konig GM, Schaberle TF |
Sci Rep |
10.1038/s41598-018-34954-y |
2018 |
* |
| Biotechnology |
Heterologous Production of the Marine Myxobacterial Antibiotic Haliangicin and Its Unnatural Analogues Generated by Engineering of the Biochemical Pathway. |
Sun Y, Feng Z, Tomura T, Suzuki A, Miyano S, Tsuge T, Mori H, Suh JW, Iizuka T, Fudou R, Ojika M |
Sci Rep |
10.1038/srep22091 |
2016 |
* |
| Metabolism |
Isolation and Biosynthetic Analysis of Haliamide, a New PKS-NRPS Hybrid Metabolite from the Marine Myxobacterium Haliangium ochraceum. |
Sun Y, Tomura T, Sato J, Iizuka T, Fudou R, Ojika M |
Molecules |
10.3390/molecules21010059 |
2016 |
* |
| Phylogeny |
A novel family VII esterase with industrial potential from compost metagenomic library. |
Kang CH, Oh KH, Lee MH, Oh TK, Kim BH, Yoon J |
Microb Cell Fact |
10.1186/1475-2859-10-41 |
2011 |
* |
| Genetics |
Investigation of cytochromes P450 in myxobacteria: excavation of cytochromes P450 from the genome of Sorangium cellulosum So ce56. |
Khatri Y, Hannemann F, Perlova O, Muller R, Bernhardt R |
FEBS Lett |
10.1016/j.febslet.2011.04.035 |
2011 |
* |
| Genetics |
Complete genome sequence of Haliangium ochraceum type strain (SMP-2). |
Ivanova N, Daum C, Lang E, Abt B, Kopitz M, Saunders E, Lapidus A, Lucas S, Glavina Del Rio T, Nolan M, Tice H, Copeland A, Cheng JF, Chen F, Bruce D, Goodwin L, Pitluck S, Mavromatis K, Pati A, Mikhailova N, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Detter JC, Brettin T, Rohde M, Goker M, Bristow J, Markowitz V, Eisen JA, Hugenholtz P, Kyrpides NC, Klenk HP |
Stand Genomic Sci |
10.4056/sigs.69.1277 |
2010 |
* |
| Cultivation |
Characteristics and living patterns of marine myxobacterial isolates. |
Zhang YQ, Li YZ, Wang B, Wu ZH, Zhang CY, Gong X, Qiu ZJ, Zhang Y |
Appl Environ Microbiol |
10.1128/AEM.71.6.3331-3336.2005 |
2005 |
* |
| Phylogeny |
Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: novel moderately halophilic myxobacteria isolated from coastal saline environments. |
Fudou R, Jojima Y, Iizuka T, Yamanaka S |
J Gen Appl Microbiol |
10.2323/jgam.48.109 |
2002 |
* |
|
 References References-
| #5321 |
Leibniz Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH ; Curators of the DSMZ;
DSM 14365
|
-
-
-
-
| #66794 |
Antje Chang, Lisa Jeske, Sandra Ulbrich, Julia Hofmann, Julia Koblitz, Ida Schomburg, Meina Neumann-Schaal, Dieter Jahn, Dietmar Schomburg:
BRENDA, the ELIXIR core data resource in 2021: new developments and updates.
Nucleic Acids Res. 49:
D498 - D508
2020 (
DOI 10.1093/nar/gkaa1025 , PubMed 33211880 )
|
-
| #67770 |
Japan Collection of Microorganism (JCM) ; Curators of the JCM;
|
-
| #69479 |
João F Matias Rodrigues, Janko Tackmann,Gregor Rot, Thomas SB Schmidt, Lukas Malfertheiner, Mihai Danaila,Marija Dmitrijeva, Daniela Gaio, Nicolas Näpflin and Christian von Mering. University of Zurich.:
MicrobeAtlas 1.0 beta
.
|
-
-
-
| #75349 |
Reimer, L.C., Lissin, A.,Schober, I., Witte,J.F., Podstawka, A., Lüken, H., Bunk, B.,Overmann, J.:
StrainInfo: A central database for resolving microbial strain identifiers
.
(
DOI 10.60712/SI-ID100667.1 )
|
- * These data were automatically processed and therefore are not curated
Change proposal
Successfully sent
|
Name and taxonomic classification

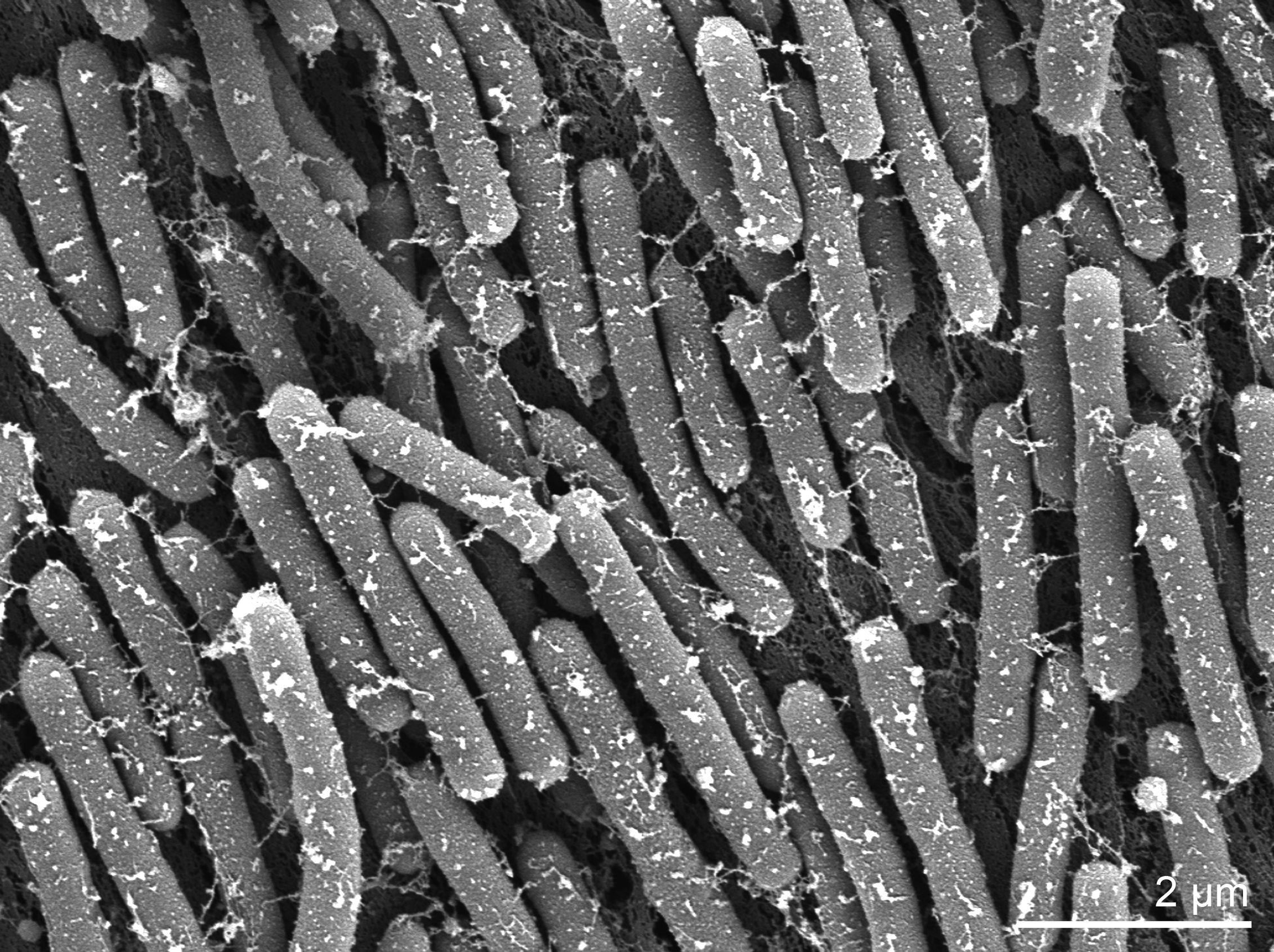
Morphology

Culture and growth conditions

Physiology and metabolism

Isolation, sampling and environmental information

Safety information

Sequence information

Genome-based predictions

External links

References