| [Ref.: #1142] |
Sample type/isolated from |
pine litter |
| [Ref.: #1142] |
Geographic location (country and/or sea, region) |
Brisbane, Alderley |
| [Ref.: #1142] |
Country |
Australia |
| [Ref.: #1142] |
Country ISO 3 Code |
AUS |
| [Ref.: #1142] |
Continent |
Australia and Oceania |
| |
| [Ref.: #43362] |
Sample type/isolated from |
from an infusion of litter from the base of a pine tree |
| [Ref.: #43362] |
Geographic location (country and/or sea, region) |
Alderley, Brisbane |
| [Ref.: #43362] |
Country |
Australia |
| [Ref.: #43362] |
Country ISO 3 Code |
AUS |
| [Ref.: #43362] |
Continent |
Australia and Oceania |
| [Ref.: #43362] |
Geographic location |
-27.0000°/153.0000° |
| |
| [Ref.: #121834] |
Sample type/isolated from |
Pine needles |
| [Ref.: #121834] |
Geographic location (country and/or sea, region) |
Adlerly, Queensland |
| [Ref.: #121834] |
Country |
Australia |
| [Ref.: #121834] |
Country ISO 3 Code |
AUS |
| [Ref.: #121834] |
Continent |
Australia and Oceania |
| [Ref.: #121834] |
Isolation date |
1979 |
|
* marker position based on {}
|
|
Isolation sources categories |
| #Host |
#Plants |
#Tree |
| #Host |
#Plants |
#Decomposing plant |
|
 |
-
Availability in culture collections
 External links External links
| [Ref.: #1142] |
Culture collection no. |
DSM 2588, ATCC 43595, IFO 15968, NBRC 15968, UQM 2034, CIP 107364, ACM 2034, NCIMB 11800 |
| [Ref.: #86730] |
 SI-ID 7622 SI-ID 7622
|
* |
 |
Literature: |
Only first 10 entries are displayed. Click here to see all.Click here to see only first 10 entries. |
|
Topic |
Title |
Authors |
Journal |
DOI |
Year |
|
| Metabolism |
Functional Analysis of a Gene Cluster from Chitinophaga pinensis Involved in Biosynthesis of the Pyrrolidine Azasugar DAB-1. |
Nunez C, Horenstein NA |
J Nat Prod |
10.1021/acs.jnatprod.9b00758 |
2019 |
* |
| Phylogeny |
Chitinophaga rhizophila sp. nov., isolated from rhizosphere soil of banana. |
Zhang XJ, Feng GD, Yang F, Zhu H, Yao Q |
Int J Syst Evol Microbiol |
10.1099/ijsem.0.005118 |
2021 |
* |
| Phylogeny |
Chitinophaga agri sp. nov., a bacterium isolated from soil of reclaimed land. |
Lee SA, Heo J, Kim TW, Sang MK, Song J, Kwon SW, Weon HY |
Arch Microbiol |
10.1007/s00203-020-02066-9 |
2020 |
* |
| Phylogeny |
In Silico Characterization of a Unique Plant-Like "Loopful" GH19 Chitinase from Newly Isolated Chitinophaga sp. YS-16. |
Sharma S, Singh R, Kaur R |
Curr Microbiol |
10.1007/s00284-020-02022-2 |
2020 |
* |
| Phylogeny |
Filobacterium rodentium gen. nov., sp. nov., a member of Filobacteriaceae fam. nov. within the phylum Bacteroidetes; includes a microaerobic filamentous bacterium isolated from specimens from diseased rodent respiratory tracts. |
Ike F, Sakamoto M, Ohkuma M, Kajita A, Matsushita S, Kokubo T |
Int J Syst Evol Microbiol |
10.1099/ijsem.0.000685 |
2015 |
* |
| Phylogeny |
Thermoflavifilum aggregans gen. nov., sp. nov., a thermophilic and slightly halophilic filamentous bacterium from the phylum Bacteroidetes. |
Anders H, Dunfield PF, Lagutin K, Houghton KM, Power JF, MacKenzie AD, Vyssotski M, Ryan JLJ, Hanssen EG, Moreau JW, Stott MB |
Int J Syst Evol Microbiol |
10.1099/ijs.0.057463-0 |
2014 |
* |
| Phylogeny |
Chitinophaga oryziterrae sp. nov., isolated from the rhizosphere soil of rice (Oryza sativa L.). |
Chung EJ, Park TS, Jeon CO, Chung YR |
Int J Syst Evol Microbiol |
10.1099/ijs.0.036442-0 |
2012 |
* |
| Genetics |
Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). |
Glavina Del Rio T, Abt B, Spring S, Lapidus A, Nolan M, Tice H, Copeland A, Cheng JF, Chen F, Bruce D, Goodwin L, Pitluck S, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Chain P, Saunders E, Detter JC, Brettin T, Rohde M, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lucas S |
Stand Genomic Sci |
10.4056/sigs.661199 |
2010 |
* |
| Phylogeny |
Chitinophaga eiseniae sp. nov., isolated from vermicompost. |
Yasir M, Chung EJ, Song GC, Bibi F, Jeon CO, Chung YR |
Int J Syst Evol Microbiol |
10.1099/ijs.0.023028-0 |
2010 |
* |
|
Fixed-target serial crystallography at the Structural Biology Center. |
Sherrell DA, Lavens A, Wilamowski M, Kim Y, Chard R, Lazarski K, Rosenbaum G, Vescovi R, Johnson JL, Akins C, Chang C, Michalska K, Babnigg G, Foster I, Joachimiak A |
J Synchrotron Radiat |
10.1107/S1600577522007895 |
2022 |
* |
|
 References References-
| #1142 |
Leibniz Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH ; Curators of the DSMZ;
DSM 2588
|
-
-
-
| #33117 |
; Curators of the CIP;
|
-
| #43362 |
Tijana Glavina Del Rio, Birte Abt, Stefan Spring, Alla Lapidus, Matt Nolan, Hope Tice, Alex Copeland, Jan-Fang Cheng, Feng Chen, David Bruce, Lynne Goodwin, Sam Pitluck, Natalia Ivanova, Konstantinos Mavromatis, Natalia Mikhailova, Amrita Pati, Amy Chen, Krishna Palaniappan, Miriam Land, Loren Hauser, Yun-Juan Chang, Cynthia D. Jeffries, Patrick Chain, Elizabeth Saunders, John C. Detter, Thomas Brettin, Manfred Rohde, Markus Göker, Jim Bristow, Jonathan A. Eisen, Victor Markowitz, Philip Hugenholtz, Nikos C. Kyrpides, Hans-Peter Klenk, Susan Lucas:
Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034).
Stand Genomic Sci 2:
87 - 95
2010 (
DOI 10.1099/00207713-31-3-285 )
|
-
-
-
-
| #66794 |
Antje Chang, Lisa Jeske, Sandra Ulbrich, Julia Hofmann, Julia Koblitz, Ida Schomburg, Meina Neumann-Schaal, Dieter Jahn, Dietmar Schomburg:
BRENDA, the ELIXIR core data resource in 2021: new developments and updates.
Nucleic Acids Res. 49:
D498 - D508
2020 (
DOI 10.1093/nar/gkaa1025 , PubMed 33211880 )
|
-
| #68382 |
Automatically annotated from API zym .
|
-
| #69479 |
João F Matias Rodrigues, Janko Tackmann,Gregor Rot, Thomas SB Schmidt, Lukas Malfertheiner, Mihai Danaila,Marija Dmitrijeva, Daniela Gaio, Nicolas Näpflin and Christian von Mering. University of Zurich.:
MicrobeAtlas 1.0 beta
.
|
-
-
-
| #86730 |
Reimer, L.C., Lissin, A.,Schober, I., Witte,J.F., Podstawka, A., Lüken, H., Bunk, B.,Overmann, J.:
StrainInfo: A central database for resolving microbial strain identifiers
.
(
DOI 10.60712/SI-ID7622.1 )
|
-
| #121834 |
Collection of Institut Pasteur ; Curators of the CIP;
CIP 107364
|
-
| #124043 |
Dr. Isabel Schober, Dr. Julia Koblitz:
Data extracted from sequence databases, automatically matched based on designation and taxonomy .
|
- * These data were automatically processed and therefore are not curated
Change proposal
Successfully sent
|
|
Name and taxonomic classification

Morphology

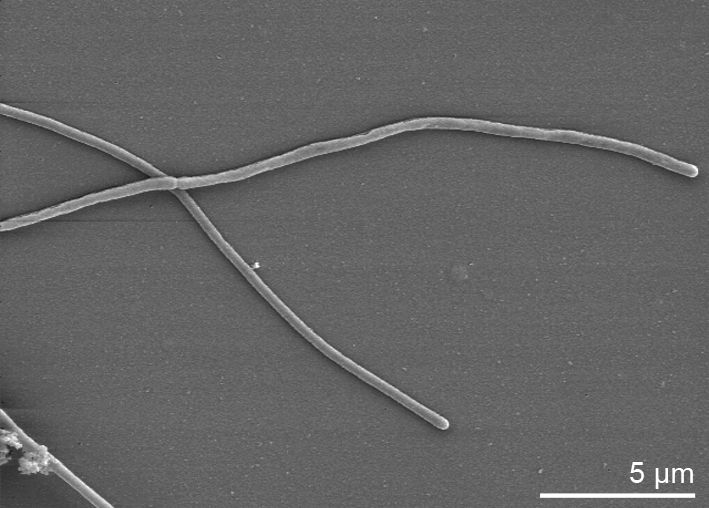
Culture and growth conditions

Physiology and metabolism

Isolation, sampling and environmental information

Safety information

Sequence information

Genome-based predictions

External links

References