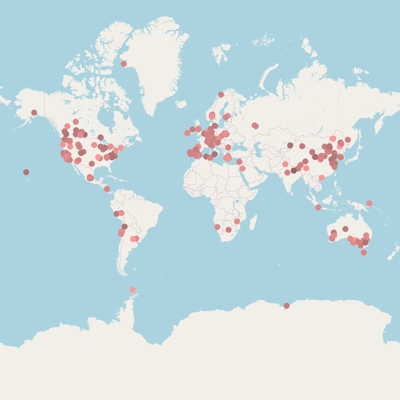
| [Ref.: #7207] |
Sample type/isolated from |
soil |
| [Ref.: #7207] |
Country |
China |
| [Ref.: #7207] |
Country ISO 3 Code |
CHN |
| [Ref.: #7207] |
Continent |
Asia |
| |
| [Ref.: #67770] |
Sample type/isolated from |
Soil |
| [Ref.: #67770] |
Geographic location (country and/or sea, region) |
near Beijing |
| [Ref.: #67770] |
Country |
China |
| [Ref.: #67770] |
Country ISO 3 Code |
CHN |
| [Ref.: #67770] |
Continent |
Asia |
| |
| [Ref.: #119374] |
Sample type/isolated from |
Environment, Soil |
| [Ref.: #119374] |
Country |
China |
| [Ref.: #119374] |
Country ISO 3 Code |
CHN |
| [Ref.: #119374] |
Continent |
Asia |
|
* marker position based on {}
|
|
Isolation sources categories |
| #Environmental |
#Terrestrial |
#Soil |
|
 |
-
Availability in culture collections
 External links External links
| [Ref.: #7207] |
Culture collection no. |
DSM 17836, IFO 14399, JCM 10339, KCTC 9580, NBRC 14399, KACC 20248, CIP 107494, IMSNU 22160, NCIMB 13631, NRRL B-16234 |
| [Ref.: #80325] |
 SI-ID 62448 SI-ID 62448
|
* |
 |
Literature: |
Only first 10 entries are displayed. Click here to see all.Click here to see only first 10 entries. |
|
Topic |
Title |
Authors |
Journal |
DOI |
Year |
|
| Metabolism |
Two Novel Glycoside Hydrolases Responsible for the Catabolism of Cyclobis-(1-->6)-alpha-nigerosyl. |
Tagami T, Miyano E, Sadahiro J, Okuyama M, Iwasaki T, Kimura A |
J Biol Chem |
10.1074/jbc.M116.727305 |
2016 |
* |
| Enzymology |
Cloning, expression and characterization of a novel GH5 exo/endoglucanase of Thermobi fi da halotolerans YIM 90462(T) by genome mining. |
Zhang F, Zhang XM, Yin YR, Li WJ |
J Biosci Bioeng |
10.1016/j.jbiosc.2015.04.012 |
2015 |
* |
| Phylogeny |
Kribbella amoyensis sp. nov., isolated from rhizosphere soil of a pharmaceutical plant, Typhonium giganteum Engl. |
Xu Z, Xu Q, Zheng Z, Huang Y |
Int J Syst Evol Microbiol |
10.1099/ijs.0.033290-0 |
2011 |
* |
| Genetics |
Complete genome sequence of Kribbella flavida type strain (IFO 14399). |
Pukall R, Lapidus A, Glavina Del Rio T, Copeland A, Tice H, Cheng JF, Lucas S, Chen F, Nolan M, Labutti K, Pati A, Ivanova N, Mavromatis K, Mikhailova N, Pitluck S, Bruce D, Goodwin L, Land M, Hauser L, Chang YJ, Jeffries CD, Chen A, Palaniappan K, Chain P, Rohde M, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Brettin T |
Stand Genomic Sci |
10.4056/sigs.731321 |
2010 |
* |
| Phylogeny |
Kribbella ginsengisoli sp. nov., isolated from soil of a ginseng field. |
Cui YS, Lee JS, Lee ST, Im WT |
Int J Syst Evol Microbiol |
10.1099/ijs.0.008516-0 |
2009 |
* |
| Phylogeny |
Kribbella catacumbae sp. nov. and Kribbella sancticallisti sp. nov., isolated from whitish-grey patinas in the catacombs of St Callistus in Rome, Italy. |
Urzi C, De Leo F, Schumann P |
Int J Syst Evol Microbiol |
10.1099/ijs.0.65613-0 |
2008 |
* |
| Phylogeny |
Kribbella karoonensis sp. nov. and Kribbella swartbergensis sp. nov., isolated from soil from the Western Cape, South Africa. |
Kirby BM, Roes ML, Meyers PR |
Int J Syst Evol Microbiol |
10.1099/ijs.0.63951-0 |
2006 |
* |
| Phylogeny |
Classification of 'Nocardioides fulvus' IFO 14399 and Nocardioides sp. ATCC 39419 in Kribbella gen. nov., as Kribbella flavida sp. nov. and Kribbella sandramycini sp. nov. |
Park YH, Yoon JH, Shin YK, Suzuki K, Kudo T, Seino A, Kim HJ, Lee JS, Lee ST |
Int J Syst Bacteriol |
10.1099/00207713-49-2-743 |
1999 |
* |
|
 References References-
| #7207 |
Leibniz Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH ; Curators of the DSMZ;
DSM 17836
|
-
-
-
| #33808 |
; Curators of the CIP;
|
-
-
-
| #66794 |
Antje Chang, Lisa Jeske, Sandra Ulbrich, Julia Hofmann, Julia Koblitz, Ida Schomburg, Meina Neumann-Schaal, Dieter Jahn, Dietmar Schomburg:
BRENDA, the ELIXIR core data resource in 2021: new developments and updates.
Nucleic Acids Res. 49:
D498 - D508
2020 (
DOI 10.1093/nar/gkaa1025 , PubMed 33211880 )
|
-
| #67770 |
Japan Collection of Microorganism (JCM) ; Curators of the JCM;
|
-
| #68371 |
Automatically annotated from API 50CH acid .
|
-
| #68382 |
Automatically annotated from API zym .
|
-
| #69479 |
João F Matias Rodrigues, Janko Tackmann,Gregor Rot, Thomas SB Schmidt, Lukas Malfertheiner, Mihai Danaila,Marija Dmitrijeva, Daniela Gaio, Nicolas Näpflin and Christian von Mering. University of Zurich.:
MicrobeAtlas 1.0 beta
.
|
-
-
| #80325 |
Reimer, L.C., Lissin, A.,Schober, I., Witte,J.F., Podstawka, A., Lüken, H., Bunk, B.,Overmann, J.:
StrainInfo: A central database for resolving microbial strain identifiers
.
(
DOI 10.60712/SI-ID62448.1 )
|
-
| #119374 |
Collection of Institut Pasteur ; Curators of the CIP;
CIP 107494
|
- * These data were automatically processed and therefore are not curated
Change proposal
Successfully sent
|
|
Name and taxonomic classification

Morphology

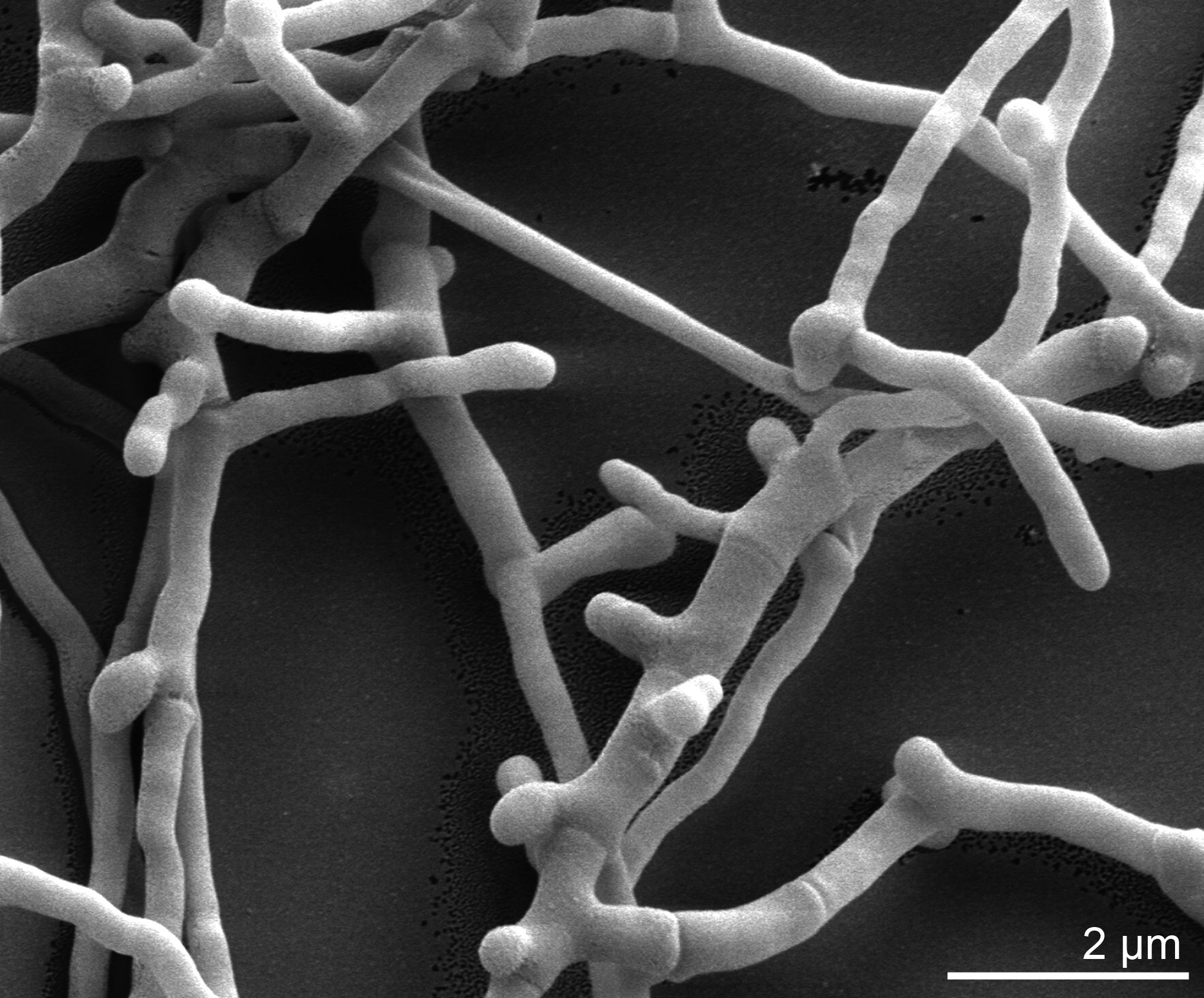
Culture and growth conditions

Physiology and metabolism

Isolation, sampling and environmental information

Safety information

Sequence information

Genome-based predictions

External links

References